Today, let’s unravel the complexities of Oligonucleotide Synthesis, with a special focus on the intricate dance of molecules in the solid-phase chemistry steps. At Biolytic Lab Performance, Inc., our commitment to advancing biotechnology is reflected in the precision and expertise embedded in every product we offer – from consumables to DNA RNA oligo synthesizers and post-synthesis accessories.
The Foundation: Designing the Oligonucleotide Sequence
Before we dive into the solid-phase chemistry intricacies, it all starts with sequence design. Whether you’re an experienced researcher or new to oligo synthesis, our consumables and synthesizers are designed to accommodate a myriad of sequences, providing flexibility and adaptability to your specific research needs.

Solid-Phase Synthesis:
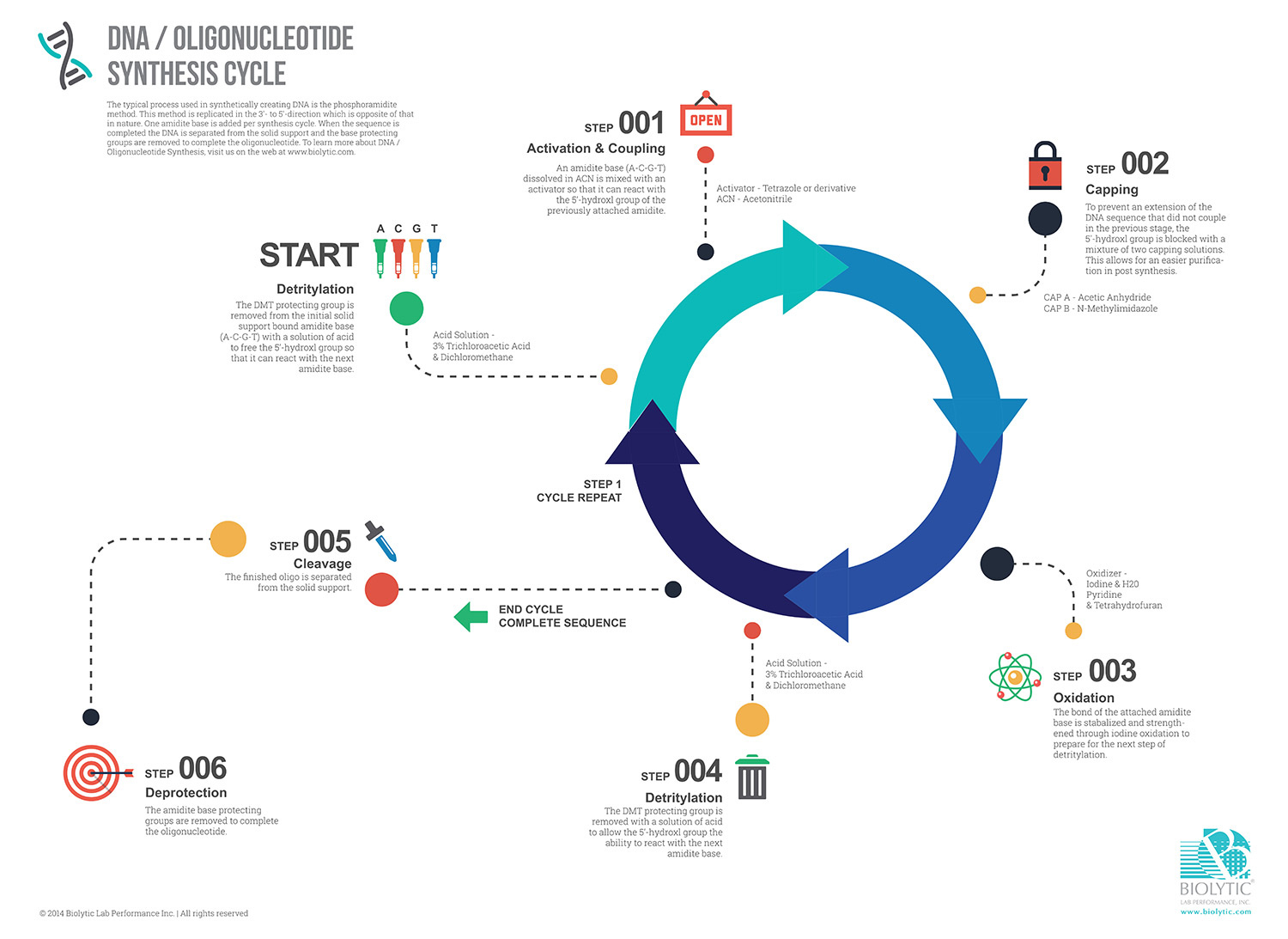
Step 1: Initiation & Detritylation/Deblocking
The process commences with the solid support, often resin-bound, acting as the canvas. The first nucleotide, protected by specific groups, is then anchored. Through controlled chemistry, the protecting groups are selectively removed, revealing the reactive sites for the subsequent steps. The DMT protecting group is removed from the initial solid support bound amidite base (A-C-G-T) with a solution of acid to free the 5’-hydroxyl group so that it can react with the next base. Acid solution typically comprises of 3% Trichloroacetic Acid and Dichloromethane.
Step 2: Activation & Coupling
In the heart of solid-phase chemistry, activation and coupling are key steps. Each nucleotide is meticulously added to the growing oligonucleotide chain. First, the amidite base is dissolved in acetonitrile (ACN) and mixed with an activator, typically Tetrazole or derivative, so that it can react with the 5’-hydroxyl group of the previously attached amidite.
Step 3: Capping
To maintain fidelity in the sequence, any unreacted or truncated sequences are capped, preventing their unwanted inclusion in the final product. The 5’-hydroxyl group is blocked with a mixture of two capping solutions, Acetic Anhydride and N-Methylimidazole. The capping process has an added benefit of allowing for easier purification in post-synthesis.
Step 4: Oxidation
Simultaneously, oxidation ensures that the newly added nucleotide is securely bonded to the growing chain. The bond is stabilized and strengthened through iodine oxidation to prepare for the next step.
Step 5: Detritylation/Deblocking
As the chain extends, protecting groups must be selectively removed to expose reactive sites for the next coupling. This deblocking step requires precision to avoid unintended reactions and to maintain the accuracy of the sequence. The DMT protecting group is removed with a solution of acid to allow the 5’-hydroxyl group the ability to react with the next amidite base.
Step 5: Repetition
The coupling, capping, oxidation, and deblocking cycle repeats until the desired oligonucleotide sequence is fully assembled.
Post-Synthesis: Nurturing the Synthesized Oligonucleotide
Step 1: Cleavage and Deprotection
Once the synthesis is complete, to use the oligo in downstream applications, the oligo must be freed from the solid support through cleavage and the protecting group removed through deprotection.
Step 2: Purification and Quality Control
Purification becomes paramount to remove any impurities or by-products. Length, purity, and sequence validation are meticulously assessed, ensuring the final product meets the highest standards.
Solid Phase Synthesis: FAQ

Elevate Your Research with Biolytic Lab Performance, Inc.
At Biolytic Lab Performance, Inc., our passion lies in empowering researchers with tools to turn ideas into oligos.
Whether you’re charting new territories in biotech research or navigating the intricacies of solid-phase chemistry, trust in Biolytic Lab Performance, Inc. to be your partner in discovery.

Explore our selection of end-to-end oligonucleotide synthesis products.